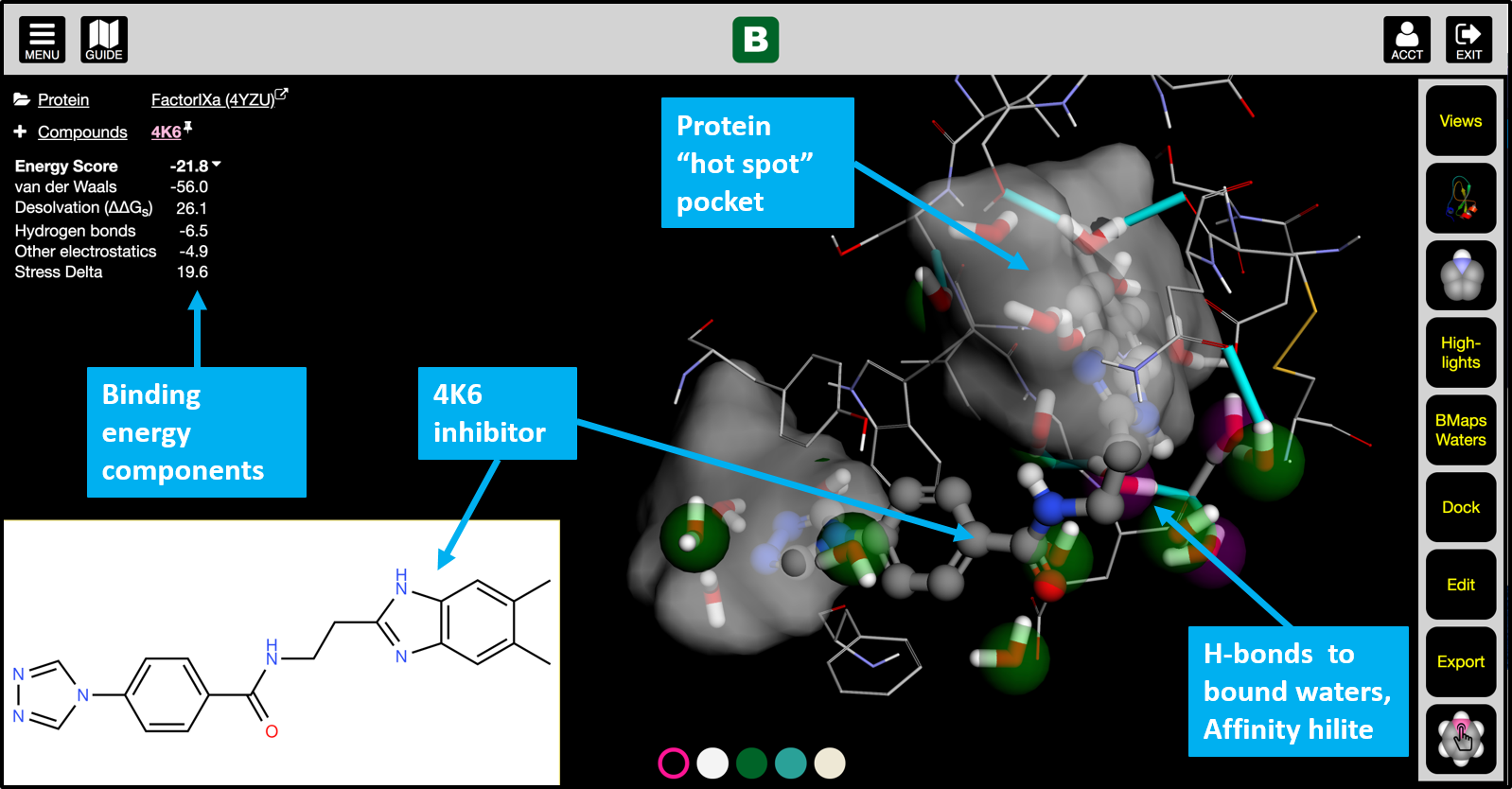
- Where does my compound bind to the protein?
- Is it binding near protein “hot spots”?
- What factors contribute to the binding and by how much?
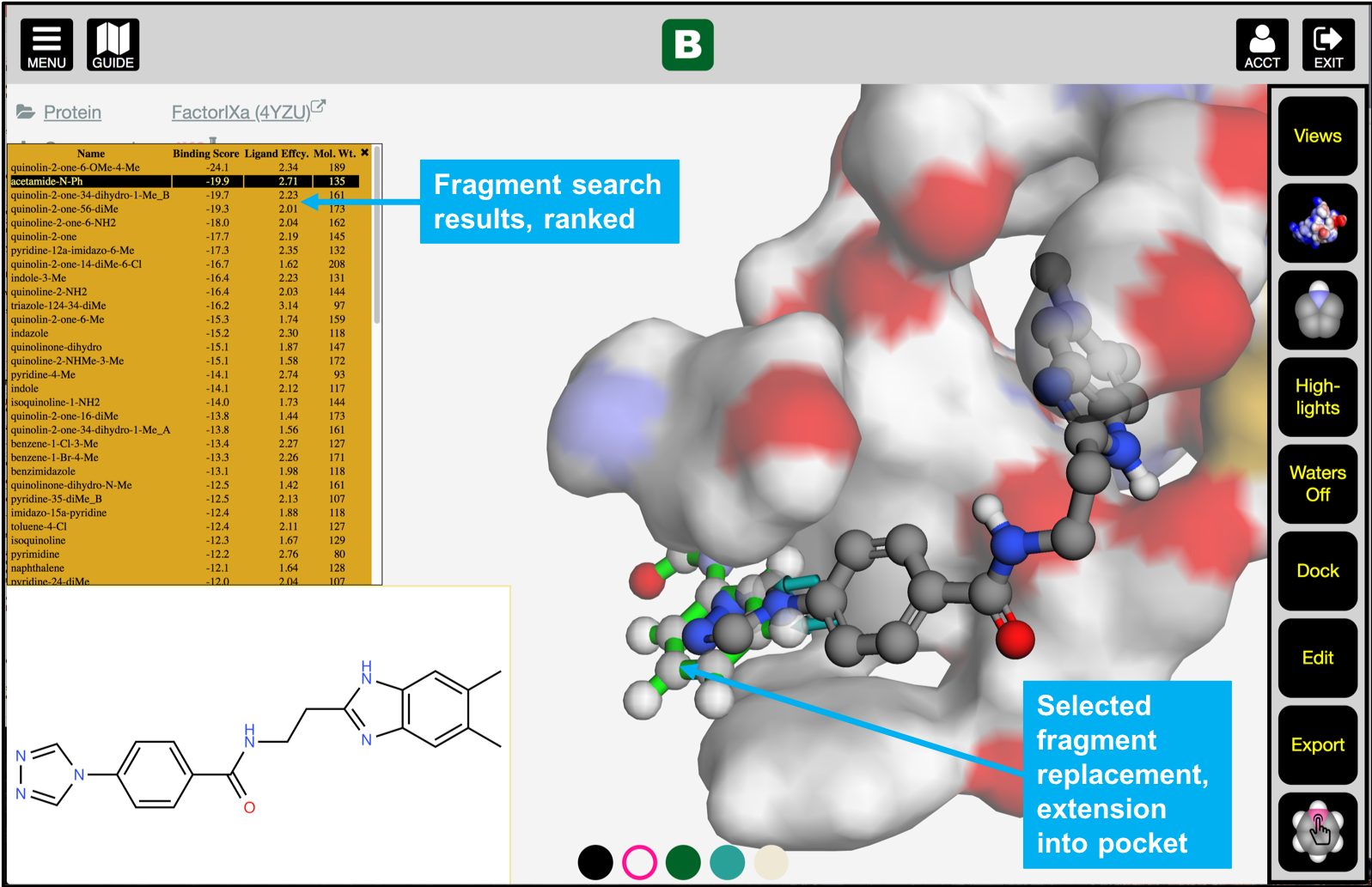
- What chemical substitutions are predicted to improve binding affinity, ligand efficiency, and
solubility?
- Where are tightly-bound waters and how do they impact ligand binding?
- Will modifications result in compounds that avoid existing patents?
- What chemical motifs lead to highly-specific binding that improves selectivity to avoid
off-target effects and mutation resistance?
- Select a protein by family, name, or PDB ID
- Drag-and-drop your compounds in any of many common formats,
or draw it with the chemical editor
- Dock compounds with a single button click to find binding poses
- Visualize interactions in the binding site
- View details of binding energy, desolvation, stress
- Modify functional groups in either the 3D or 2D views
- Search fragment maps for potent compound modifications,
ranked by binding affinity
- Export designed compounds to molecule files,
SMILES strings, or various Web services

